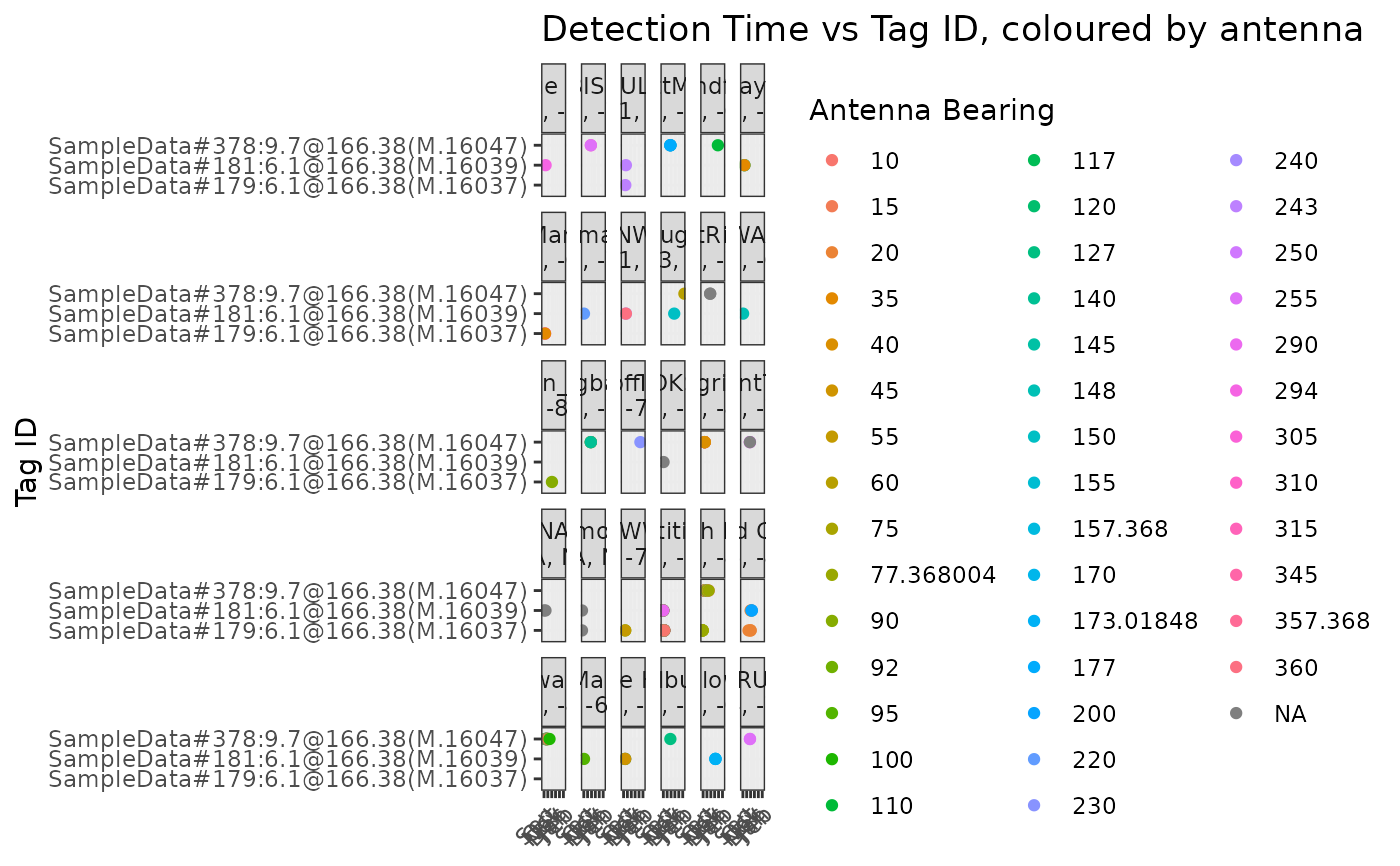
Plot tag ID vs time for all tags detected by site, coloured by antenna
bearing. Input is expected to be a data frame, database table, or database.
The data must contain "ts", "antBearing", "fullID", "recvDeployName",
"recvDeployLat", "recvDeployLon", and optionally "gpsLat" and "gpsLon". If
GPS lat/lon are included, they will be used rather than recvDeployLat/Lon.
These data are generally contained in the alltags or the alltagsGPS
views. If a motus database is submitted, the alltagsGPS view will be used.
Arguments
- df_src
Data frame, SQLite connection, or SQLite table. An SQLite connection would be the result of
tagme(XXX)orDBI::dbConnect(RSQLite::SQLite(), "XXX.motus"); an SQLite table would be the result ofdplyr::tbl(tags, "alltags"); a data frame could be the result ofdplyr::tbl(tags, "alltags") %>% dplyr::collect().- sitename
Character vector. Subset of sites to plot. If
NULL, all unique sites are plotted.- ncol
Numeric. Passed on to
ggplot2::facet_wrap()- nrow
Numeric. Passed on to
ggplot2::facet_wrap()- data
Defunct, use
src,df_src, ordfinstead.
Examples
# Download sample project 176 to .motus database (username/password are "motus.sample")
if (FALSE) sql_motus <- tagme(176, new = TRUE) # \dontrun{}
# Or use example data base in memory
sql_motus <- tagmeSample()
# convert sql file "sql_motus" to a tbl called "tbl_alltags"
library(dplyr)
tbl_alltags <- tbl(sql_motus, "alltagsGPS")
# Plot all sites within file for tbl file tbl_alltags
plotSite(tbl_alltags)
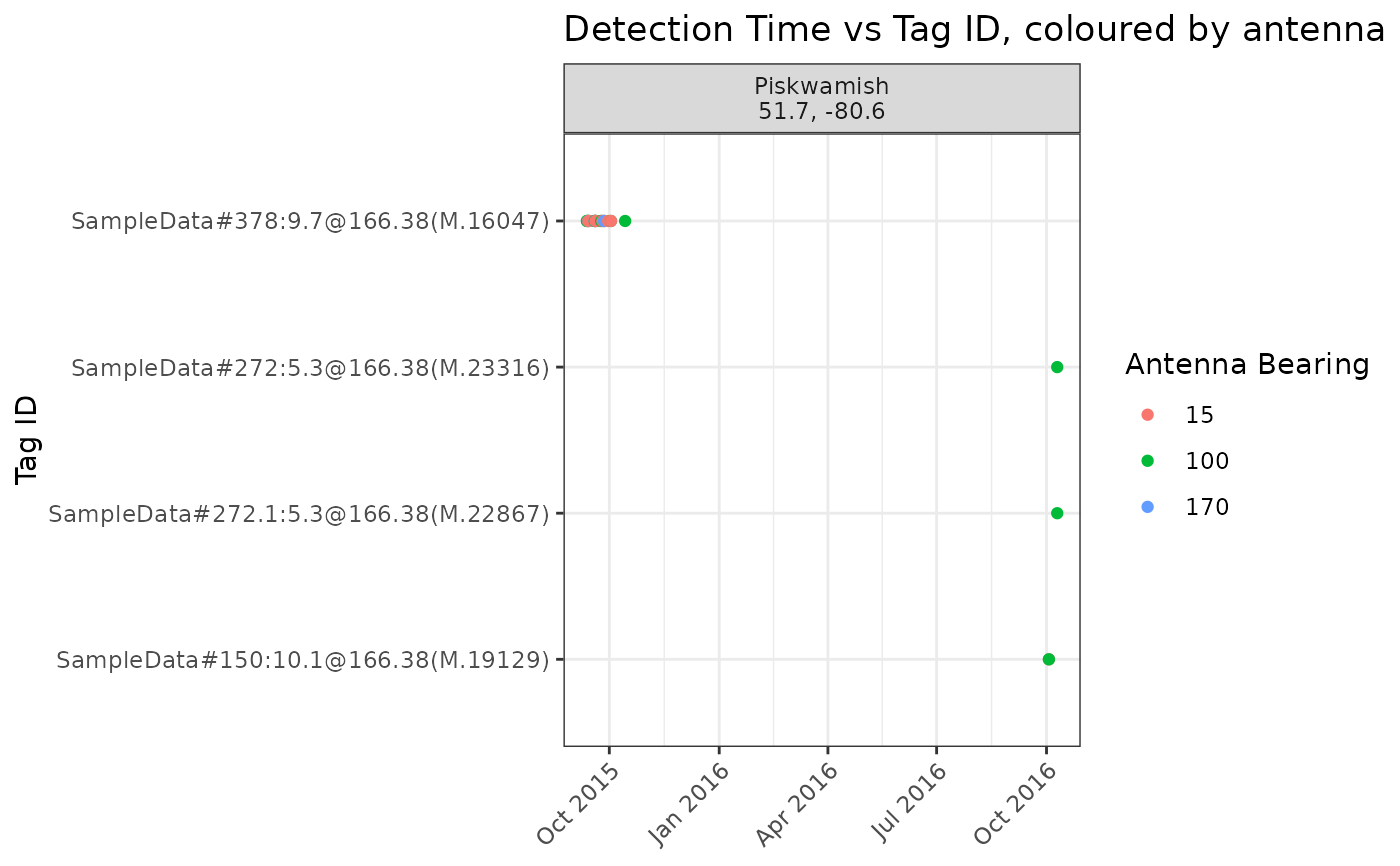
 # Plot only detections at a specific site; Piskwamish
plotSite(tbl_alltags, sitename = "Piskwamish")
# Plot only detections at a specific site; Piskwamish
plotSite(tbl_alltags, sitename = "Piskwamish")
 # For more custom filtering, convert the tbl "tbl_alltags" to a data.frame called "df_alltags"
df_alltags <- collect(tbl_alltags)
# Plot only detections for specified tags for data.frame df_alltags
plotSite(filter(df_alltags, motusTagID %in% c(16047, 16037, 16039)))
# For more custom filtering, convert the tbl "tbl_alltags" to a data.frame called "df_alltags"
df_alltags <- collect(tbl_alltags)
# Plot only detections for specified tags for data.frame df_alltags
plotSite(filter(df_alltags, motusTagID %in% c(16047, 16037, 16039)))